The MVTec 3D-AD Dataset for Unsupervised 3D Anomaly Detection and Localization#
Authors: Paul Bergmann, Xin Jin, David Sattlegger, Carsten Steger
Affiliations: MVTec Software GmbH, Technical University of Munich, Karsruhe Institute of Technology
Links: Project Page, arXiv
Summary#
The authors introduce the first comprehensive 3D dataset for the task of unsupervised anomaly detection and localization. A high-resolution industrial 3D sensor is used to acquire depth scans of 10 different object categories.
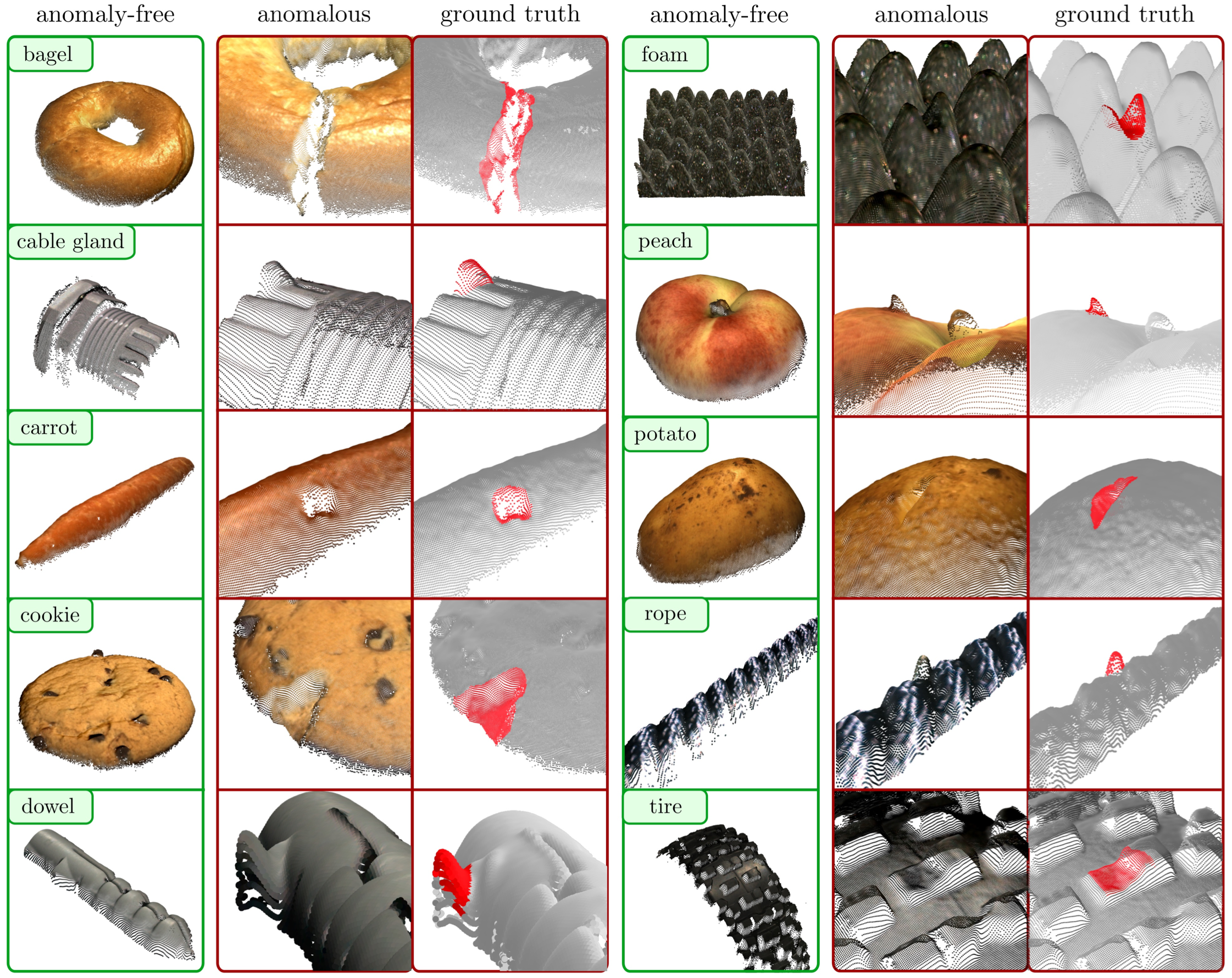
Key Ideas#
The MVTec 3D-AD dataset. The dataset consists of 4147 scans acquired by a high-resolution industrial 3D sensor. For each category, a set of anomaly-free scans is provided for model training and validation.
Performance evaluation. The model output a real-valued anomaly score for each \((x, y, z)\) pixel of the test set, which is then converted to binary predictions using a threshold. Then we compute the per-region overlap (PRO) metric, which is defined as the average relative overlap of the binary prediction \(P\) with each connected component \(C_k\) of the ground truth.
This process is repeated for multiple thresholds and a curve is constructed by plotting the resulting PRO values against the corresponding false positive rates. The final performance is computed by integrating this curve up to a limited false positive rate (i.e., 0.3) and normalising the resulting area to the interval \([0, 1]\).
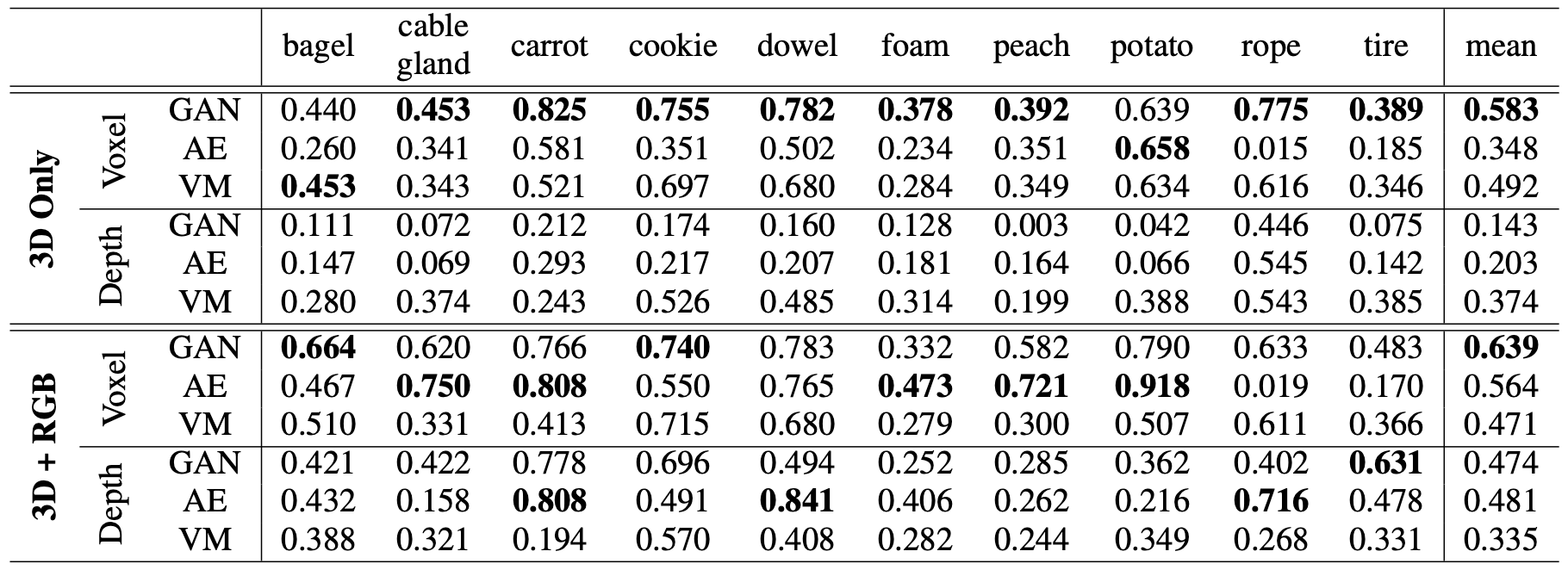
Initial benchmark. Very few methods have been proposed explicitly for this task and all of them operate on voxel data. The authors consider Voxel f-AnoGAN [1] and their implementation of a convolutional Voxel AE [2].
Technical Details#
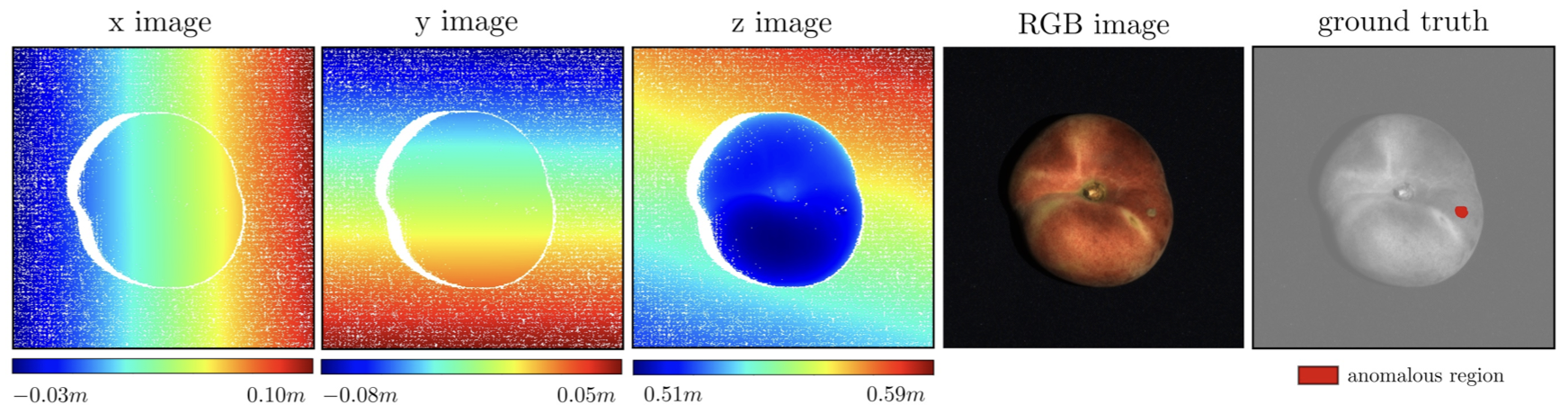
Visualization of the provided data for one anomalous test sample of the dataset category peach.
Anomaly localization results.
Notes#
References#
[1] Simarro Viana, Jaime, Ezequiel de la Rosa, Thijs Vande Vyvere, David Robben, and Diana M. Sima. “Unsupervised 3d brain anomaly detection.” In International MICCAI Brainlesion Workshop, pp. 133-142. Springer, Cham, 2020.
[2] Bengs, Marcel, Finn Behrendt, Julia Krüger, Roland Opfer, and Alexander Schlaefer. “Three-dimensional deep learning with spatial erasing for unsupervised anomaly segmentation in brain MRI.” International journal of computer assisted radiology and surgery 16, no. 9 (2021): 1413-1423.